

- #Snapgene viewer uppercase copy paste how to
- #Snapgene viewer uppercase copy paste update
- #Snapgene viewer uppercase copy paste full
However, different programs may use the EDITSEQ file type for different types of data.
#Snapgene viewer uppercase copy paste update
We are continually working on adding more file type descriptions to the site, so if you have information about EDITSEQ files that you think will help others, please use the Update Info link below to submit it to us - we'd love to hear from you! While we do not yet describe the EDITSEQ file format and its common uses, we do know which programs are known to open these files, as we receive dozens of suggestions from users like yourself every day about specific file types and which programs they use to open them. Important: Different programs may use files with the EDITSEQ file extension for different purposes, so unless you are sure which format your EDITSEQ file is, you may need to try a few different programs. While we have not verified the app ourselves yet, our users have suggested a single EDITSEQ opener which you will find listed below.SnapGene Crack Free Download is an easy-to-use genome browser that supports a variety of file formats. SnapGene has two modes of operation: the "Map" and the "Sequence" modes. Map mode displays restriction sites along with gene annotations on the map as well as on the sequence. In sequence mode, users can annotate the sequence and place restriction sites by dragging and dropping the user-defined areas on the sequence. They can also create and edit restriction sites. In Map mode, users can manipulate the map either directly on the screen or in a designated "Map Panel" window. In the map panel, the sequence can be viewed and zoomed, the map can be rotated or scaled, annotations can be moved and copied, and additional features such as primers, restriction sites, etc.
#Snapgene viewer uppercase copy paste how to
This demo shows how to prepare a PCR product and use it to isolate the template DNA. This example is designed for use with the GeneMapper 3.7.2 software, or later versions of GeneMapper. GeneMapper 3.7.2 or later is required to view this example.
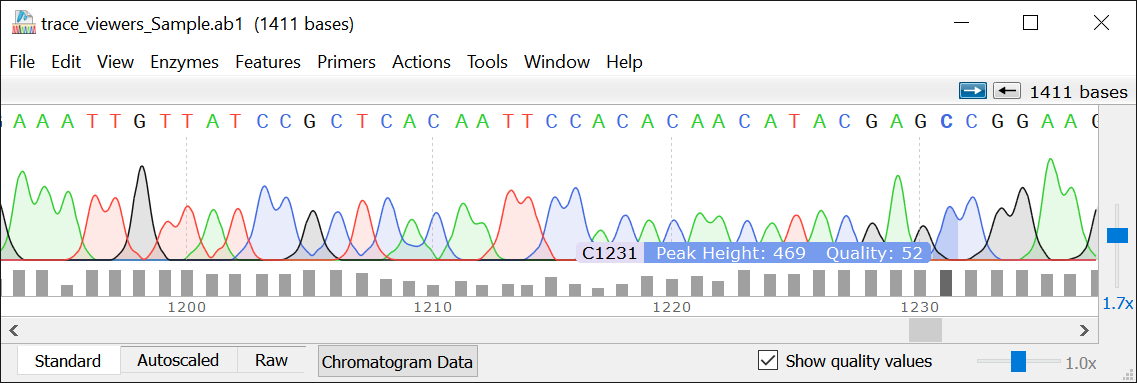
GeneMapper 3.7.2 or later is required for viewing this example. To prepare the template DNA, use a 200- to 500-bp PCR product as a template. Note: The PCR product should be purified before use. If purification is not performed, the template may not produce the desired results.ġ. Select Copy and Paste from the Cut menu.Ģ. Select the Copy Mode option on the menu.ģ. Select Paste and choose Copy from the Paste menu.Ĥ. In the Target field, enter either the following text or the template name.ĥ. In the Format menu, select Size from the Selection tab.Ħ. Click the Size menu item and select the Size tab.Ĩ. Click OK twice to exit from the window.ġ. Select the Size drop-down menu and choose Size.ģ. Click OK twice to exit from the window.Ģ.
#Snapgene viewer uppercase copy paste full
(See the accompanying file COPYING.LIB for full copyright information) Select the Band Type option and choose Single Strand or Double Strand. is a versatile, easy-to-use and feature-rich program that allows you to manipulate gene sequences. It provides a convenient graphical interface for the management and display of DNA fragments.

enables you to store, compare, edit and visualize gene sequences and restriction maps. * Manipulating nucleotide and amino acid sequences Moreover, it provides a unique facility for detecting the presence of restriction sites in the DNA fragments.


 0 kommentar(er)
0 kommentar(er)
